Proteome Browser
The CGD Proteome Browser
interactively displays conserved domains identified using the protein sequence to query the
InterPro database.
The Domains/Motifs page displays a thumbnail image of the browser; clicking on that brings
up the browser itself.
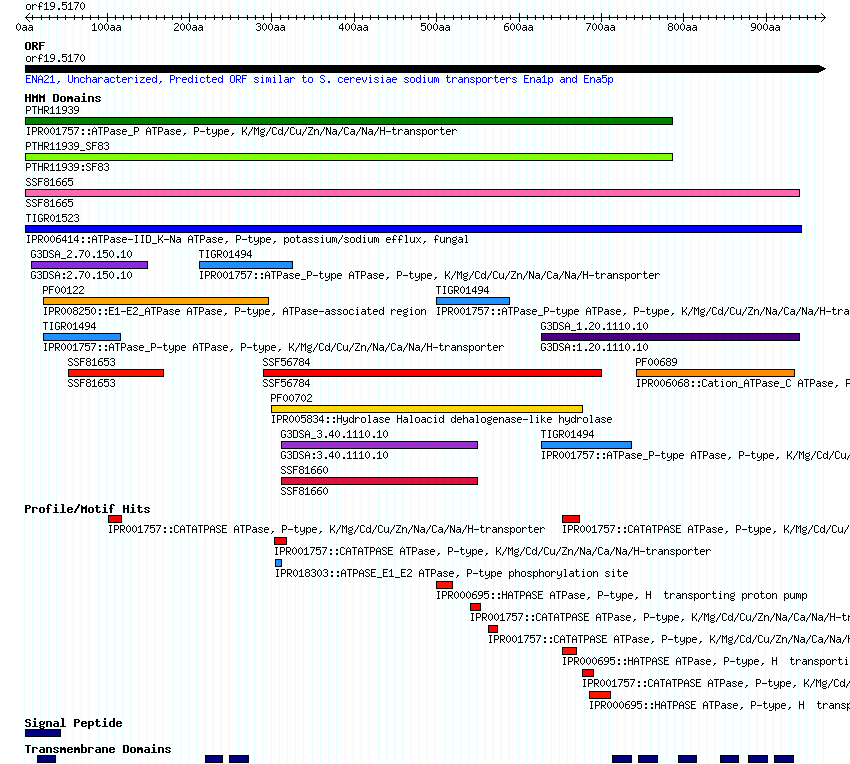
Shared Domains/Motifs
Table providing information about other proteins (in the same organism) that also contain features identified in the query protein sequence. The table has three columns:
- Protein. The name of the protein sharing features with the query, with a link to its Locus Summary page.
- Features in common with query protein. List of InterPro domains or motifs identified in both query protein and this protein. Includes the source database, the feature accession number (linked to a description of the feature), and a brief description of the feature.
- Other features in this protein (but not in query protein). List of InterPro domains or motifs identified in this protein but absent in query. Includes the source database, the feature accession number (linked to a description of the feature), and a brief description of the feature.
Unique Domains/Motifs
Table listing features found in the query protein, but which are found in no other other proteins in the organism. The table has three columns:
- Database source. InterPro member database where the feature was defined.
- Accession number. Feature identifier, linked to a description of the feature at the source database.
- Description Brief description of the name and function of the feature, if known.
Transmembrane Domains
Table showing transmembrane domains (if any) identified in the query protein using the TMHMM program. The table has two columns:
- Predicted Transmembrane Domain(s). A Proteome Browser thumbnail image showing the relative position in the protein of transmembrane domains. Clicking on the image opens a browser session.
- Amino Acid Coordinates. A table listing the start and end positions of each transmembrane domain.
Signal Peptides
Table showing signal peptides (if any) identified in the query protein using the SignalP program. The table has two columns:
- Predicted Signal Peptide(s). A Proteome Browser thumbnail image showing the relative position in the protein of signal peptides. Clicking on the image opens a browser session.
- Amino Acid Coordinates. A table listing the start and end positions of each signal peptide.
External Links
For your convenience, we also provide several links for searching external resources with the query sequence. Depending on your specific problem or interest, one or the other of these may produce the best results for you. For specific help with any of these other resources, you will be best served at the original source. Currently we support searches of the following resources:
 Return to CGD
Return to CGD