"Search" section

- Landmark or Region. Find a different protein for display. Search by gene name, systematic name or keyword.
- Reports & Analysis.
Display or download a summary of the page information by clicking "Go". Change
default format or display options by clicking "Configure...". Options include:
- Download Decorated FASTA File. Optionally configure which features (domains, motifs etc.) to highlight in the sequence.
- Download GFF File. Download page information as General Feature Format (GFF) file for use on your local system.
- Download Sequence File. A number of formats are available through configuration.
- Scroll/Zoom. Control the views in the Overview and Details windows. Arrow buttons control the position, and zoom is controlled with the pull-down menu or +/- buttons. The default zoom is the full-length protein.
"Overview" section

Graduated scale showing full length of protein. Red box indicates the region of the protein shown in Details window, controlled by Scroll/Zoom option in the Search section. Clicking on the scale re-centers the red box/Details window.
"Details" section
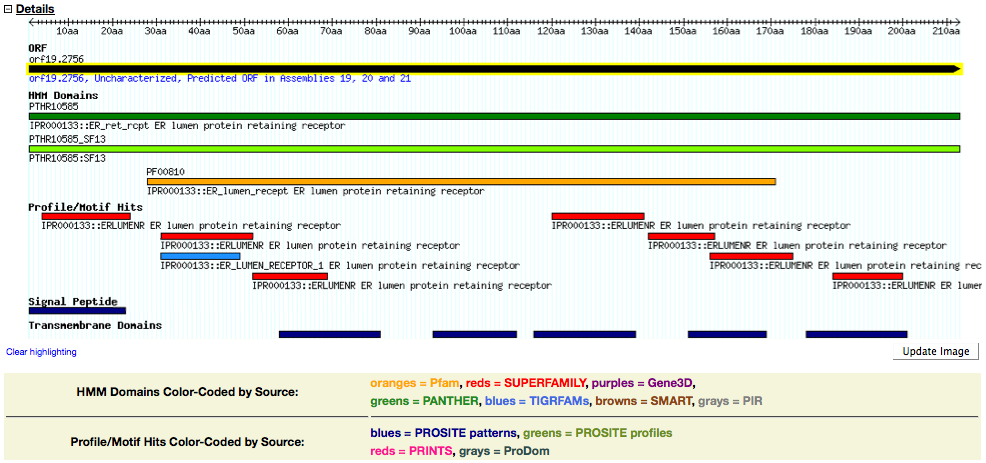
Presents feature information in tracks along the primary structure of the protein. The lower four tracks display features detected by searching the protein sequence using the InterProScan program.
- ORF. Track representing the full-length protein. Clicking on the track opens the CGD Locus Summary page for the encoding gene.
- HMM Domains. Matches to domains defined on the basis of hidden Markov models (HMMs) built from protein families at the member databases comprising InterPro:
- Profile/Motif Hits. Matches to specific sequence patterns or sequence alignment profiles, defined at several InterPro member databases:
- Signal Peptide. Matches to signal peptide signatures detected with the SignalP search algorithm.
- Transmembrane Domains. Matches to transmembrane domain signatures detected with the TMHMM search algorithm.
"Tracks" section

Allows user control over which tracks are displayed n the Details window, as well as their display order.
"Display Settings" section
Allows user control over the appearance of the Details window, including size, labeling and highlighting.
"Add your own tracks" section
Allows the addition of user-defined tracks in the Details window, either via an uploaded file or remote URL.
 Return to CGD
Return to CGD