Candida Strains These lists and lineage diagrams have been compiled by CGD curators to serve as a brief reference to some of the more commonly used laboratory strains of various Candida species, rather than a comprehensive resource. The strain names on each species list below link to short summary descriptions, notes and citations. If you would like to suggest additions, corrections, or updates to this list, please send a message to CGD curators with details. |
 |
||
|
|||
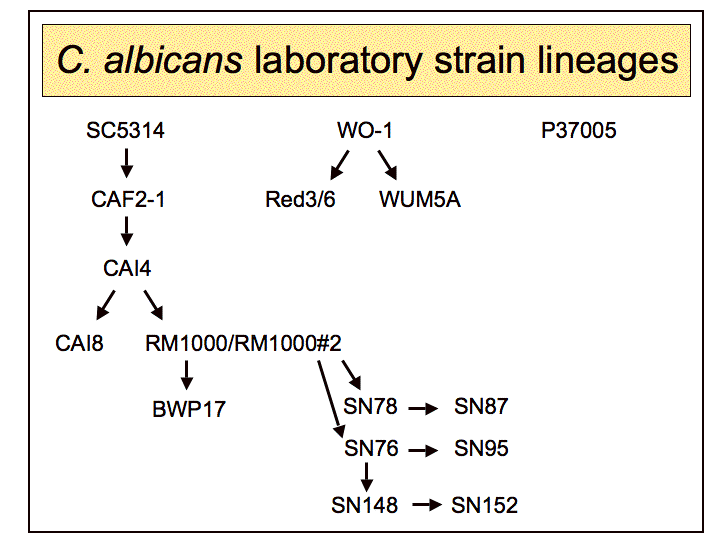 |
| SC5314 | ||
| Genotype: Wild-type | ||
| Notes: Wild-type strain used in the systematic sequencing project, the reference sequence stored in CGD. The original strain background from which most of the common laboratory strains are derived. This strain is virulent in a mouse model of systemic infection and is frequently used as a wild-type control.
In their 2004 Genome Biology paper on C. albicans genome sequence, Frank Odds, Al Brown and Neil Gow explain the origins of SC5314: "Strain SC5314 was used in the 1980s by scientists at the E.R. Squibb company* (now Bristol-Myers Squibb) for their pioneering studies of C. albicans molecular biology. It was engineered by Fonzi and Irwin to provide the uridine autotrophic mutant that has been essential to most subsequent molecular genetic research into C. albicans. The strain is usually described merely as a 'clinical isolate', but it is worth setting on record that SC5314 was originally isolated from a patient with generalized Candida infection by Margarita Silva-Hutner** at the Department of Dermatology, Columbia College of Physicians and Surgeons (New York, USA). The original isolate number was 1775 and the strain is identical with strain NYOH#4657 in the New York State Department of Health collection. (This information was provided by Joan Fung-Tome at Bristol-Myers Squibb as a personal communication.) SC5314 belongs to the predominant clade of closely related C. albicans strains that represents almost 40% of all isolates worldwide, as determined by DNA fingerprinting and multi-locus sequence typing (A. Tavanti, A.D. Davidson, N.A.R.G., M.C.J. Maiden and F.C.O., unpublished observations)." | ||
Odds FC, Brown AJ, Gow NA. Candida albicans genome sequence: a platform for genomics in the absence of genetics. Genome Biol. 2004; 5(7): 230. 

Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993 Jul;134(3):717-28. 

| ||
*The earliest publications that used SC5314 came in 1968 from Squibb Institute for Medical Research: Aszalos A, Robison RS, Lemanski P, Berk B. Trienine, an antitumor triene antibiotic. J Antibiot (Tokyo). 1968 Oct;21(10):611-5. 

Maestrone G, Semar R. Establishment and treatment of cutaneous Candida albicans infection in the rabbit. Naturwissenschaften. 1968 Feb;55(2):87-8. 

Meyers E, Miraglia GJ, Smith DA, Basch HI, Pansy FE, Trejo WH, Donovick R. Biological characterization of prasinomycin, a phosphorus-containing antibiotic. Appl Microbiol. 1968 Apr;16(4):603-8. 

| ||
**The documentation of the work done by Margarita Silva-Hutner and her lab is preserved at Columbia University Archival Collections. Back to top | ||
| CAF2-1 | ||
| Genotype: URA3/ura3::imm434 IRO1/iro1::imm434 | ||
| Notes: URA3 heterozygous strain derived from the SC5314 strain. The 3-prime end of one copy of the IRO1 gene that resides adjacent to URA3 was inadvertently deleted during the construction of this strain. This strain is virulent in a mouse model of systemic infection and is frequently used as a wild-type control. | ||
Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993 Jul;134(3):717-28. 

Back to top | ||
| CAI4 | ||
| Genotype: ura3::imm434/ura3::imm434 iro1/iro1::imm434 | ||
| Notes: Isogenic to the SC5314 strain. Uridine auxotroph constructed by deletion of the second copy of URA3. The second copy of IRO1 was inadvertently deleted upon strain construction. As a result, the strain and its descendants have no functional copy of IRO1. This strain is avirulent in a mouse model of systemic infection unless complemented with URA3. | ||
Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993 Jul;134(3):717-28. 

| ||
Garcia MG, O'Connor JE, Garcia LL, Martinez SI, Herrero E, del Castillo Agudo L. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast. 2001 Mar 15;18(4):301-11. 

Back to top | ||
| CAI8 | ||
| Genotype: ura3::imm434/ura3::imm434 iro1/iro1::imm434 ade2::hisG/ade2::hisG | ||
| Notes: Isogenic to the SC5314 strain. Derived from the CAF2-1 strain by deletion of URA3 and both copies of ADE2 using the URA-blaster method. | ||
Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993 Jul;134(3):717-28. 

Back to top | ||
| P37005 | ||
| Genotype: MTLa/MTLa | ||
| Notes: Wild-type clinical isolate. Naturally homozygous for the MTLa mating type locus. | ||
Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002 Oct;162(2):737-45. 

Back to top | ||
| Red3/6 | ||
| Genotype: ade2/ade2 | ||
| Notes: Isogenic to the WO-1 strain. Adenine auxotroph derived from the WO-1 strain by chemical mutagenesis using MNNG. | ||
Srikantha T, Chandrasekhar A, Soll DR. Functional analysis of the promoter of the phase-specific WH11 gene of Candida albicans. Mol Cell Biol. 1995 Mar;15(3):1797-805. 

Back to top | ||
| RM1000 | ||
| Genotype: ura3::imm434/ura3::imm434 iro1/iro1::imm434 his1::hisG/his1::hisG | ||
| Notes: Isogenic to the SC5314 strain. Derived from the CAI4 strain by deletion of the HIS1 gene using the URA-blaster method (see Fonzi and Irwin, 1993 for details of this method). The standard RM1000 strain was found to have a heterozygous deletion on chromosome 5. RM1000#2 is an isolate that has been shown to have wild-type copies of chromosome 5. | ||
Negredo A, Monteoliva L, Gil C, Pla J, Nombela C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology. 1997 Feb;143 ( Pt 2):297-302. 

Back to top | ||
| BWP17 | ||
| Genotype: ura3::imm434/ura3::imm434 iro1/iro1::imm434 his1::hisG/his1::hisG arg4/arg4 | ||
| Notes: Isogenic to the SC5314 strain. Uridine, histidine and arginine auxotroph derived from the RM1000 strain by deletion of the ARG4 gene. This strain has a heterozygous deletion on chromosome 5 that was inherited from the RM1000 parental strain. | ||
Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999 Mar;181(6):1868-74. 

Back to top | ||
| SN87 | ||
| Genotype: ura3::imm434::URA3/ura3::imm434 iro1::IRO1/iro1::imm434 his1::hisG/his1::hisG leu2/leu2 | ||
| Notes: Isogenic to the SC5314 strain. Histidine and leucine auxotroph derived from the RM1000#2 strain by deletion of the LEU2 gene. This strain is virulent in a mouse model of systemic infection. | ||
Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005 Feb;4(2):298-309. 

Back to top | ||
| SN95 | ||
| Genotype: ura3::imm434::URA3/ura3iro1IRO1/iro1his1his1arg4/arg4 | ||
| Notes: Isogenic to the SC5314 strain. Histidine and arginine auxotroph derived from the RM1000#2 strain by deletion of the ARG4 gene. This strain is virulent in a mouse model of systemic infection. | ||
Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005 Feb;4(2):298-309. 

Back to top | ||
| SN152 | ||
| Genotype: ura3/::imm434::URA3/ura3::imm434 iro1::IRO1/iro1::imm434 his1::hisG/his1::hisG leu2/leu2 arg4/arg4 | ||
| Notes: Isogenic to the SC5314 strain. Histidine, leucine and arginine auxotroph derived from the RM1000#2 strain by deletion of the LEU2 and ARG4 genes. This strain is virulent in a mouse model of systemic infection. | ||
Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005 Feb;4(2):298-309. 

Back to top | ||
| WO-1 | ||
| Genotype: MTLalpha | ||
| Notes: Wild-type clinical isolate that switches between white and opaque phenotypes at high frequency. The MTLa locus is absent in this strain (for more information see Lockhart, 2002). This strain has been sequenced by the Broad Institute (http://www.broadinstitute.org/annotation/genome/candida_group/GenomeDescriptions.html) | ||
Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. "White-opaque transition": a second high-frequency switching system in Candida albicans. J Bacteriol. 1987 Jan;169(1):189-97. 

| ||
Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002 Oct;162(2):737-45. 

| ||
| WUM5A | ||
| Genotype: MTLalpha/MTLalpha ura3-1Δ::FRT/ura3-2Δ::FRT | ||
| Notes: Isogenic to the WO-1 strain. Uridine auxotroph constructed by deletion of both copies of URA3. | ||
Strauss A, Michel S, Morschhauser J. Analysis of phase-specific gene expression at the single-cell level in the white-opaque switching system of Candida albicans. J Bacteriol. 2001 Jun;183(12):3761-9. 

Back to top | ||
Abbey D, Hickman M, Gresham D, Berman J. High-Resolution SNP/CGH Microarrays Reveal the Accumulation of Loss of Heterozygosity in Commonly Used Candida albicans Strains. G3 (Bethesda). 2011 Dec;1(7):523-30. 

Back to top | ||
 |
C. glabrata laboratory strain descriptions and references |
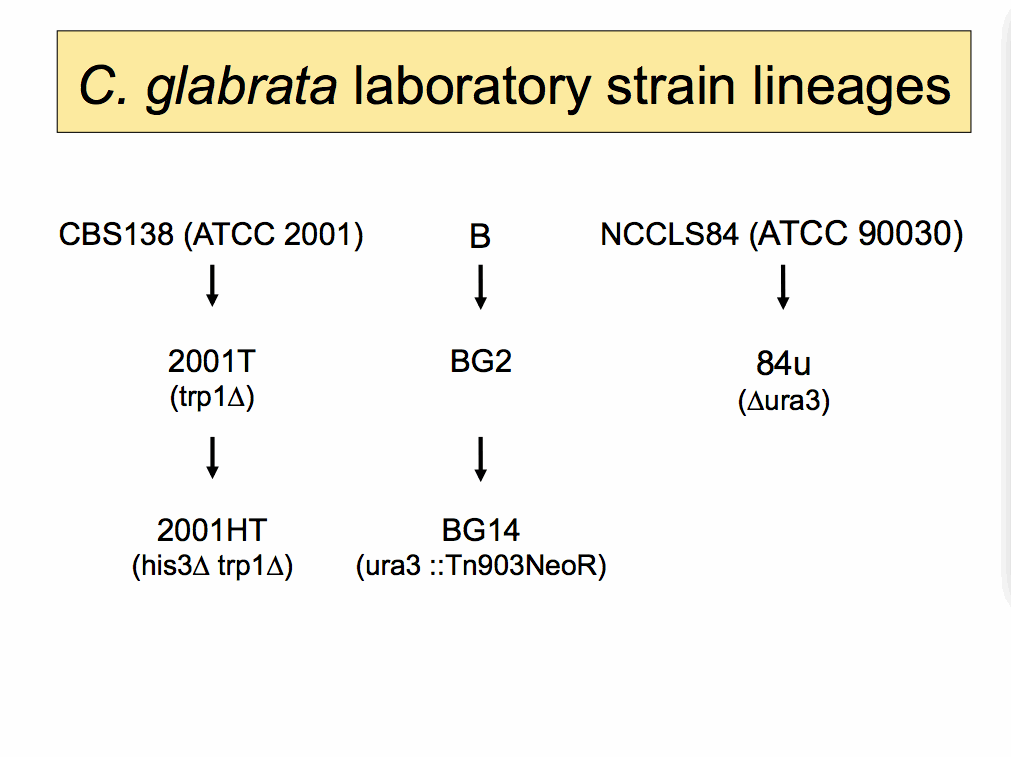
| B | ||
| Genotype: Wild-type | ||
| Notes: Cinical isolate from a case of vaginitis that did not respond to fluconazole or boric acid treatment; it is virulent in a murine model of vaginitis. This strain was called LF 547.92 in the original publication. | ||
Fidel PL Jr, Cutright JL, Tait L, Sobel JD. A murine model of Candida glabrata vaginitis. J Infect Dis. 1996 Feb;173(2):425-31. 

Back to top | ||
| BG2 | ||
| Genotype: Wild-type | ||
| Notes: Cinical isolate from a case of vaginitis that did not respond to fluconazole or boric acid treatment; it is virulent in a murine model of vaginitis. | ||
Cormack BP, Falkow S. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics. 1999 Mar;151(3):979-87. 

Back to top | ||
| BG14 | ||
| Genotype: ura3-delta(-285 +932)::Tn903NeoR | ||
| Notes: A ura3 derivitive of the wild-type B2 clinical isolate. | ||
Cormack BP, Falkow S. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics. 1999 Mar;151(3):979-87. 

Back to top | ||
| CBS138 (ATCC 2001) | ||
| Genotype: Wild-type | ||
| Notes: The reference strain for the Genolevures C. glabrata sequencing project. This strain has also been called NRRL-Y-65 and IFO 0622. | ||
Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004 Jul 1;430(6995):35-44. 

| ||
Koszul R, Malpertuy A, Frangeul L, Bouchier C, Wincker P, Thierry A, Duthoy S, Ferris S, Hennequin C, Dujon B. The complete mitochondrial genome sequence of the pathogenic yeast Candida (Torulopsis) glabrata. FEBS Lett. 2003 Jan 16;534(1-3):39-48. 

Back to top | ||
| 2001T | ||
| Genotype: trp1 | ||
| Notes: A trp1 auxotrophic strain derived from the CBS138 (ATCC 2001) wild type strain. | ||
Kitada K, Yamaguchi E, Arisawa M. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene. 1995 Nov 20;165(2):203-6. 

Back to top | ||
| 2001HT | ||
| Genotype: his3 trp1::ScURA3 | ||
| Notes:A his3 trp1 auxotrophic strain derived from 200T, a derivative of the CBS138 (ATCC 2001) wild type strain. | ||
Kitada K, Yamaguchi E, Arisawa M. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene. 1995 Nov 20;165(2):203-6. 

Back to top | ||
| NCCLS84 (ATCC90030) | ||
| Genotype: Wild-type | ||
| Notes: Wild-type strain | ||
Espinel-Ingroff A, Kish CW Jr, Kerkering TM, Fromtling RA, Bartizal K, Galgiani JN, Villareal K, Pfaller MA, Gerarden T, Rinaldi MG, et al. Collaborative comparison of broth macrodilution and microdilution antifungal susceptibility tests. J Clin Microbiol. 1992 Dec;30(12):3138-45. 

Back to top | ||
| 84u | ||
| Genotype: ura3 | ||
| Notes: A ura3 derivitive of the wild-type NCCLS84 (ATCC90030) strain. | ||
Tsai HF, Krol AA, Sarti KE, Bennett JE. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob Agents Chemother. 2006 Apr;50(4):1384-92. 

Back to top | ||
 | ||
| CDC 317 | ||
| Genotype: Wild-type | ||
| Notes: Reference strain used for the C. parapsilosis sequencing project. This isolate came from the hands of a hospital worker, who was the source for an outbreak of infection in a Mississippi community hospital in 2001. | ||
Kuhn DM, Mikherjee PK, Clark TA, Pujol C, Chandra J, Hajjeh RA, Warnock DW, Soil DR, Ghannoum MA. Candida parapsilosis characterization in an outbreak setting. Emerg Infect Dis. 2004 Jun;10(6):1074-81. 

| ||
Clark TA, Slavinski SA, Morgan J, Lott T, Arthington-Skaggs BA, Brandt ME, Webb RM, Currier M, Flowers RH, Fridkin SK, Hajjeh RA. Epidemiologic and molecular characterization of an outbreak of Candida parapsilosis bloodstream infections in a community hospital. J Clin Microbiol. 2004 Oct;42(10):4468-72. 

Back to top | ||
| GA1 | ||
| Genotype: Wild-type | ||
| Notes: Wild-type clinical isolate. The SAT1 flipper method and the Candida albicans IMH3 gene have been used as dominant-selectable markers to consruct gene knock-outs in this strain. | ||
Gacser A, Salomon S, Schafer W. Direct transformation of a clinical isolate of Candida parapsilosis using a dominant selection marker. FEMS Microbiol Lett. 2005 Apr 1;245(1):117-21. 

| ||
Gacser A, Trofa D, Schafer W, Nosanchuk JD. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J Clin Invest. 2007 Oct;117(10):3049-58. 

Back to top | ||
| CLIB214 = CBS 604 = ATCC 22019 = NRRL Y-12969 | ||
| Notes: This strain is dependent on oxidative metabolism for growth since it lacks a fermentative pathway. | ||
Ashford Bailey K (1928) Certain conditions of the gastro-intestinal tract in Porto Rico and their relation to tropical sprue. Am J Trop Med 8:507Ð538.  Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP (2011) Candida Berkhout (1923).  In: Kurtzman CP, Fell JW, Boekhout T (eds), The yeasts: a taxonomic study. p. 987-1278, 5th ed. Elsevier, London, United Kingdom.  Logue ME, Wong S, Wolfe KH, Butler G (2005) A genome sequence survey shows that the pathogenic yeast Candida parapsilosis has a defective MTLa1 allele at its mating type locus. Eukaryot Cell 4(6):1009-17. 
| ||
| SR23 = CBS7157 | ||
Kovac L, Lazowska J, Slonimski PP (1984) A yeast with linear molecules of mitochondrial DNA. Mol Gen Genet 197:420Ð424  Nosek J, Novotna M, Hlavatovicova Z, Ussery DW, Fajkus J, Tomaska L. (2004) Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol Genet Genomics 272(2):173-80.  Back to top | ||
 | ||
C. tropicalis laboratory strain descriptions and references |
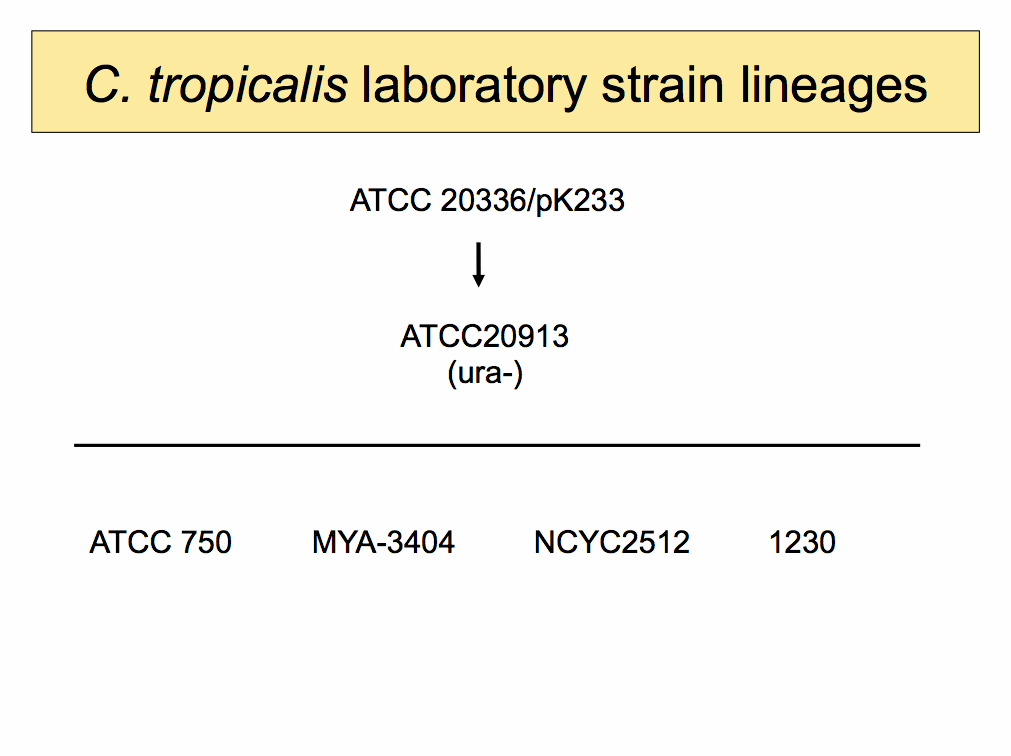
| ATCC 20336 (pK233) | |
| Genotype: Wild-type | |
| Notes: This strain excretes alpha,omega-dicarboxylic acids as a by-product when cultured on n-alkanes or fatty acids as the carbon source. | |
Craft DL, Madduri KM, Eshoo M, Wilson CR (2003) Identification and characterization of the CYP52 family of Candida tropicalis ATCC 20336, important for the conversion of fatty acids and alkanes to alpha,omega-dicarboxylic acids. Appl Environ Microbiol 69(10):5983-91. 

Back to top | |
| ATCC 20913 | |
| Genotype: ura3/ura3 | |
| Notes: A uracil auxotroph derived from C. tropicalis ATCC 20336 by random mutagenesis. | |
Haas LO, Cregg JM, Gleeson MA (1990) Development of an integrative DNA transformation system for the yeast Candida tropicalis. J Bacteriol 172(8):4571-7. 

Back to top | |
| ATCC 750 | |
| Genotype: Wild-type | |
| Notes: A wild-type fluconazole susceptible strain.. | |
Barchiesi F, Calabrese D, Sanglard D, Falconi Di Francesco L, Caselli F, Giannini D, Giacometti A, Gavaudan S, Scalise G (2000) Experimental induction of fluconazole resistance in Candida tropicalis ATCC 750. Antimicrob Agents Chemother 44(6):1578-84. 

Back to top | |
| NCYC2512 | |
| Genotype: Wild-type | |
| Notes: A wild-type saline soil isolate from Pakistan that is capable of producing large amounts of alpha,omega-dodecanedioic acid. | |
Rodriguez PL, Ali R, Serrano R (1996) CtCdc55p and CtHa13p: two putative regulatory proteins from Candida tropicalis with long acidic domains. Yeast 12(13):1321-9 

Back to top | |
| 1230 | |
| Genotype: Wild-type | |
| Notes: A wild-type dicarboxylic acid-producing industrial strain. | |
He F, Chen YT (2005) Cloning and heterologous expression of the NADPH cytochrome P450 oxidoreductase genes from an industrial dicarboxylic acid-producing Candida tropicalis. Yeast 22(6):481-91 

Back to top | |
 Return to CGD
Return to CGD